Motivation of canonical correlation analysis (CCA)
The main motivation of CCA is to provide multi-view of the data, which usually consists of two sets of variables. Canonical correlation analysis determines a set of canonical variates, i.e., orthogonal linear combinations of the variables within each set that best explain the variability both within and between sets. So there could be multiple dimensions (canonical variables) that represent the association between two sets of variables. That yields multiple aspects, aka, multi-view, for the data.
Problem setup
For
The objective (1) is equivalent to
Here we propose a common strategy in solving optimization problem (1). Denote
Extending to multiple canonical pair
Usually we only need the first several canonical pairs to view the data. So CCA is basically another way to accomplish dimension reduction.
Toy example
Let’s check it with a toy example:
A researcher has collected data on three psychological variables, four academic variables (standardized test scores) and gender for 600 college freshman. She is interested in how the set of psychological variables relates to the academic variables and gender. In particular, the researcher is interested in how many dimensions (canonical variables) are necessary to understand the association between the two sets of variables. (ref)
First, we use the existing R package CCA to conduct the analysis.
mm <- read.csv("https://stats.idre.ucla.edu/stat/data/mmreg.csv")
colnames(mm) <- c("Control", "Concept", "Motivation", "Read", "Write", "Math", "Science", "Sex")
# i.e. X: 600x3; Y: 600x5
psych <- mm[, 1:3]
acad <- mm[, 4:8]
cc1 <- cc(psych, acad)
round(cc1$xcoef,4) # i.e. U: 3xmin(3,5)## [,1] [,2] [,3]
## Control -1.2538 -0.6215 -0.6617
## Concept 0.3513 -1.1877 0.8267
## Motivation -1.2624 2.0273 2.0002round(cc1$ycoef,4) # i.e. V: 5xmin(3,5)## [,1] [,2] [,3]
## Read -0.0446 -0.0049 0.0214
## Write -0.0359 0.0421 0.0913
## Math -0.0234 0.0042 0.0094
## Science -0.0050 -0.0852 -0.1098
## Sex -0.6321 1.0846 -1.7946cc1$cor # canonical correlations## [1] 0.4640861 0.1675092 0.1039911We can also do it manually using the expression in (3).
# do it manually
mm = apply(mm, 2, function(x) scale(x, center = T, scale = F))
psych <- mm[, 1:3]
acad <- mm[, 4:8]
Sig_x = t(psych)%*%as.matrix(psych); Sig_xsqrt = sqrtm(Sig_x)
Sig_y = t(acad)%*%as.matrix(acad); Sig_ysqrt = sqrtm(Sig_y)
Sig_xy= t(psych)%*%as.matrix(acad)
MM = solve(Sig_xsqrt)%*%Sig_xy%*%solve(Sig_ysqrt)
res = svd(MM)
res$d ## which is just cc1$cor## [1] 0.4640861 0.1675092 0.1039911solve(Sig_xsqrt)%*%res$u # U## [,1] [,2] [,3]
## [1,] -0.05123026 -0.02539288 -0.02703590
## [2,] 0.01435577 -0.04852756 0.03377890
## [3,] -0.05158110 0.08283177 0.08172711solve(Sig_ysqrt)%*%res$v # V## [,1] [,2] [,3]
## [1,] -0.0018231483 -0.0002006181 0.0008735867
## [2,] -0.0014658991 0.0017189940 0.0037307163
## [3,] -0.0009568002 0.0001728118 0.0003839993
## [4,] -0.0002053221 -0.0034796325 -0.0044877370
## [5,] -0.0258276917 0.0443172840 -0.0733272900Notice that the results are the same to that from the package, except there is a scale difference.
We can construct the first canonical pair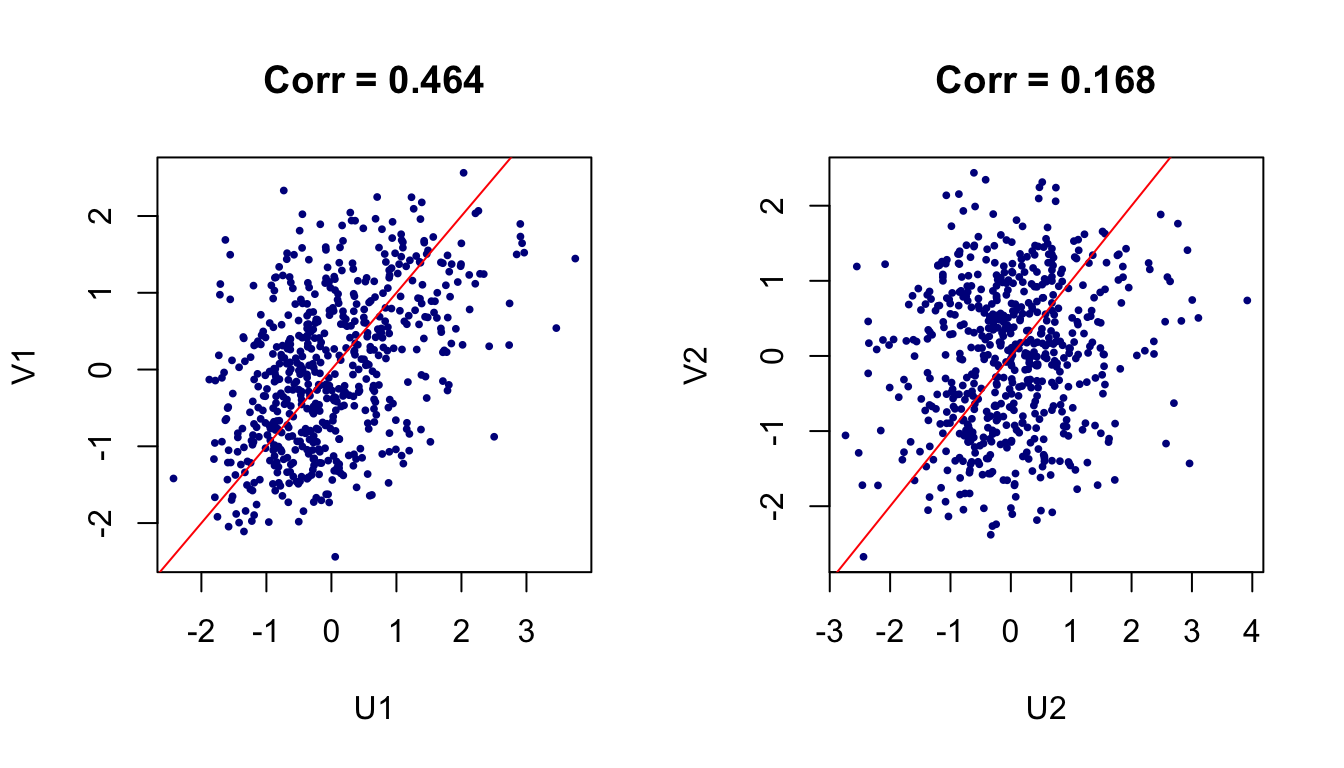
Figure 1: Viewing data on the first two canonical pairs.
Regularized CCA
For high dimensional data, it is more desirable to incorporate sparsity to construct canonical pairs using limited number of covariates. It helps to have a more clear picture of the results. Generally, we can add penalty terms to the objective function to accomplish this task, such as ridge penalty
which is equivalent to
Subgroup Identification
In previous section, we discuss about the sparsity in terms of the covariates. Another very interesting topic is to identify subgroups that are similar and easier to be described by certain canonical pairs. It is related to subgroup clustering techniques and has practical importance in many fields, like genetic studies.
Recall that the
The optimization of objective function (4) could be accomplished by coordinate descent, i.e. first take gradient descent on
Let’s see how it work through the following codes. We just conduct 10 loops and check the results. Here we pre-specified the penalty coefficient
# objective function (negative for minimization)
opt_func <- function(w) {
-cor(w*U1, w*V1) + lambda*sum(w*log(w))
}
# equality constraint
equal_fun <- function(w) {
sum(w)
}
n = dim(mm)[1]
w0 = rep(1/n,n); lambda = 1/abs(sum(w0*log(w0))) # just choose a proper lambda as an example
# let's try several rounds using coordinate descent optimization strategy
for (i in 1:10) {
cc1 <- cc(diag(w0)%*%psych, diag(w0)%*%acad) # fix w
U1 = psych %*% cc1$xcoef[,1]
V1 = acad %*% cc1$ycoef[,1]
# then fix U,V
opt_res = suppressWarnings(Rsolnp::solnp(rep(1/n,n),
opt_func,
eqfun = equal_fun,
eqB = 1,
LB = rep(1e-10,n),
UB = rep(1-1e-10,n),
control = list(trace = 0))
)
w0 = opt_res$pars
print(paste0("Objective function value: ",-round(min(opt_res$values), 4), " (Loop", i,")"))
}## [1] "Objective function value: 1.9481 (Loop1)"
## [1] "Objective function value: 1.9505 (Loop2)"
## [1] "Objective function value: 1.9511 (Loop3)"
## [1] "Objective function value: 1.9513 (Loop4)"
## [1] "Objective function value: 1.9513 (Loop5)"
## [1] "Objective function value: 1.9513 (Loop6)"
## [1] "Objective function value: 1.9513 (Loop7)"
## [1] "Objective function value: 1.9513 (Loop8)"
## [1] "Objective function value: 1.9513 (Loop9)"
## [1] "Objective function value: 1.9513 (Loop10)"The value of objective function keeps increasing and converges within 10 loops. The optimal correlation between first weighted canonical pair
However, the issue is the correlation can be greatly influenced by the outlier. Even though we penalize on the entropy, a large weight is assigned to a single point that makes the correlation quite large (see the figure below).
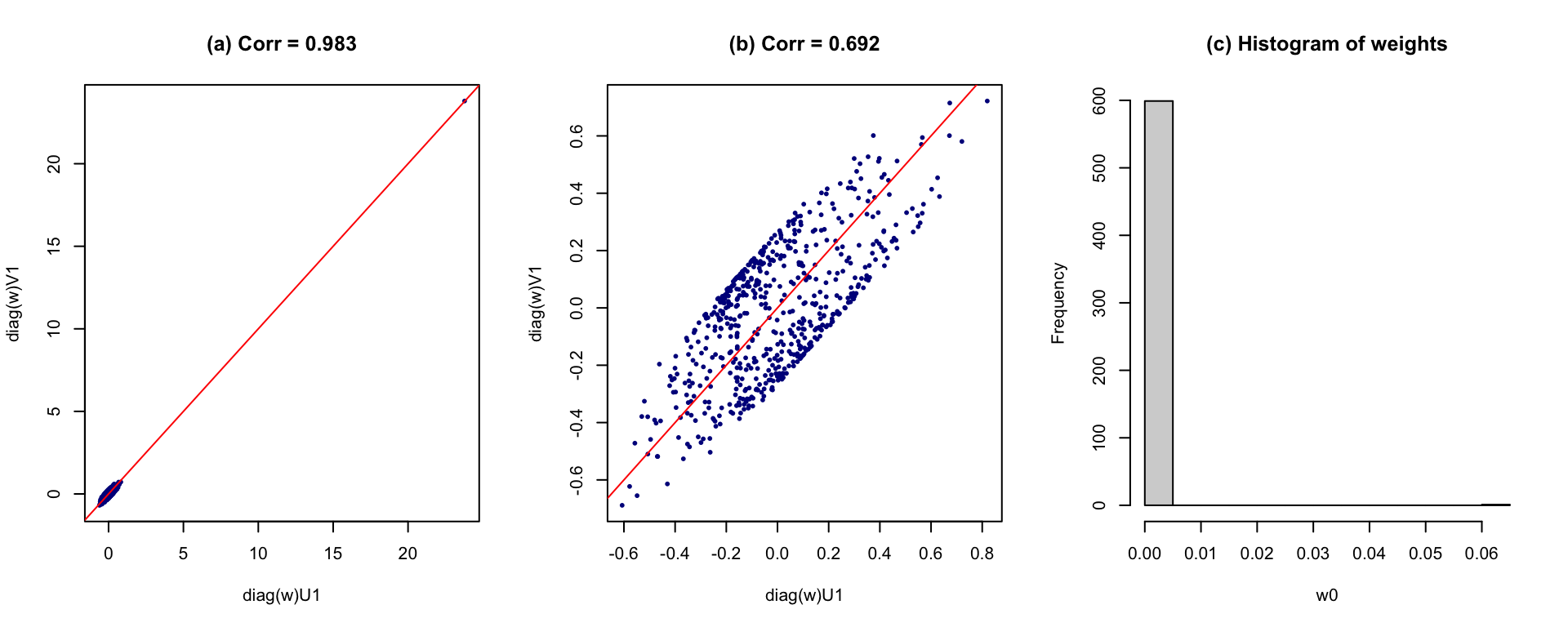
Figure 2: Results after weighting on subjects. (a) All data; (b) removing the influencial point; (c) histogram of weights
One way to handle this issue is to adopt a different penalty term that are more sensitive to the influential point. One potential proposal could be
## [1] "Objective function value: 0.8839 (Loop1)"
## [1] "Objective function value: 0.8796 (Loop2)"
## [1] "Objective function value: 0.8888 (Loop3)"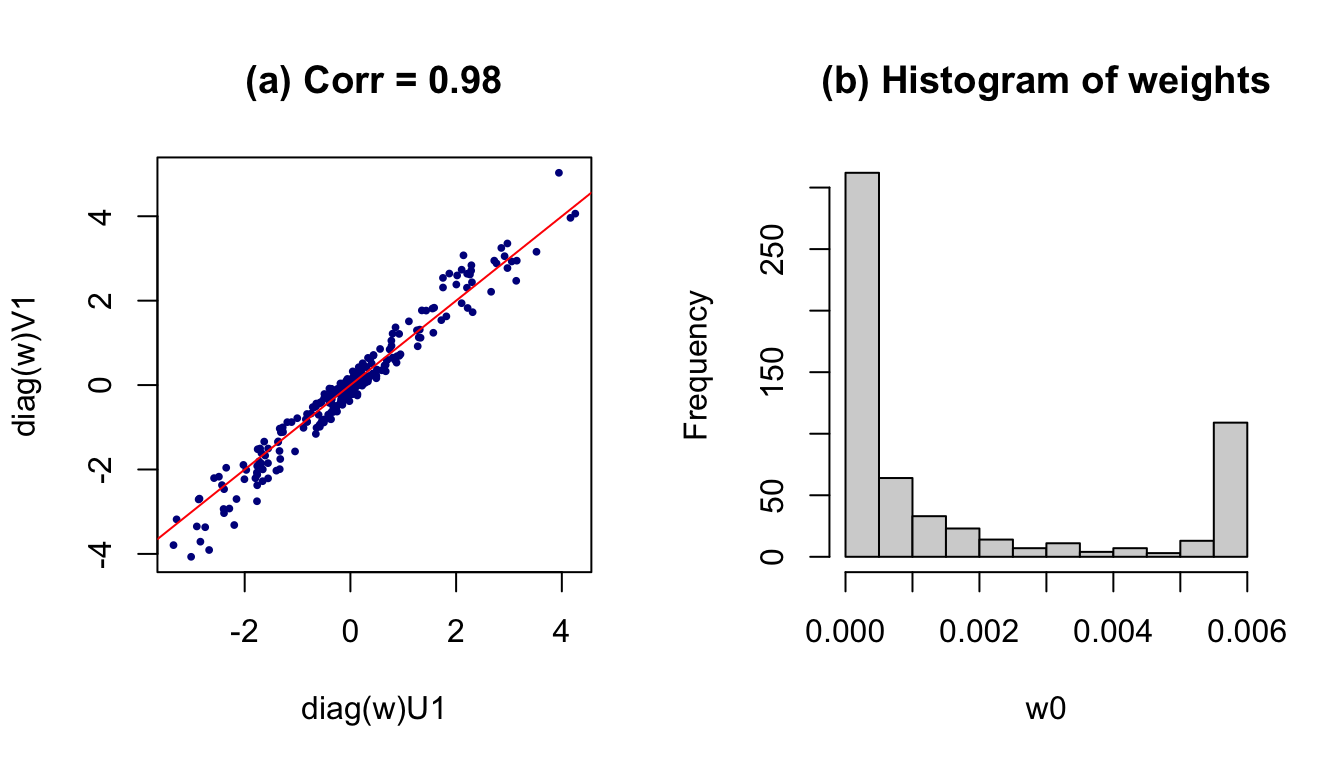
Figure 3: Results after weighting on subjects. (a) All data; (b) Histogram of weights