A corresponding Meta-learner R-package can be found on Github by the same author.
1 A General Framework
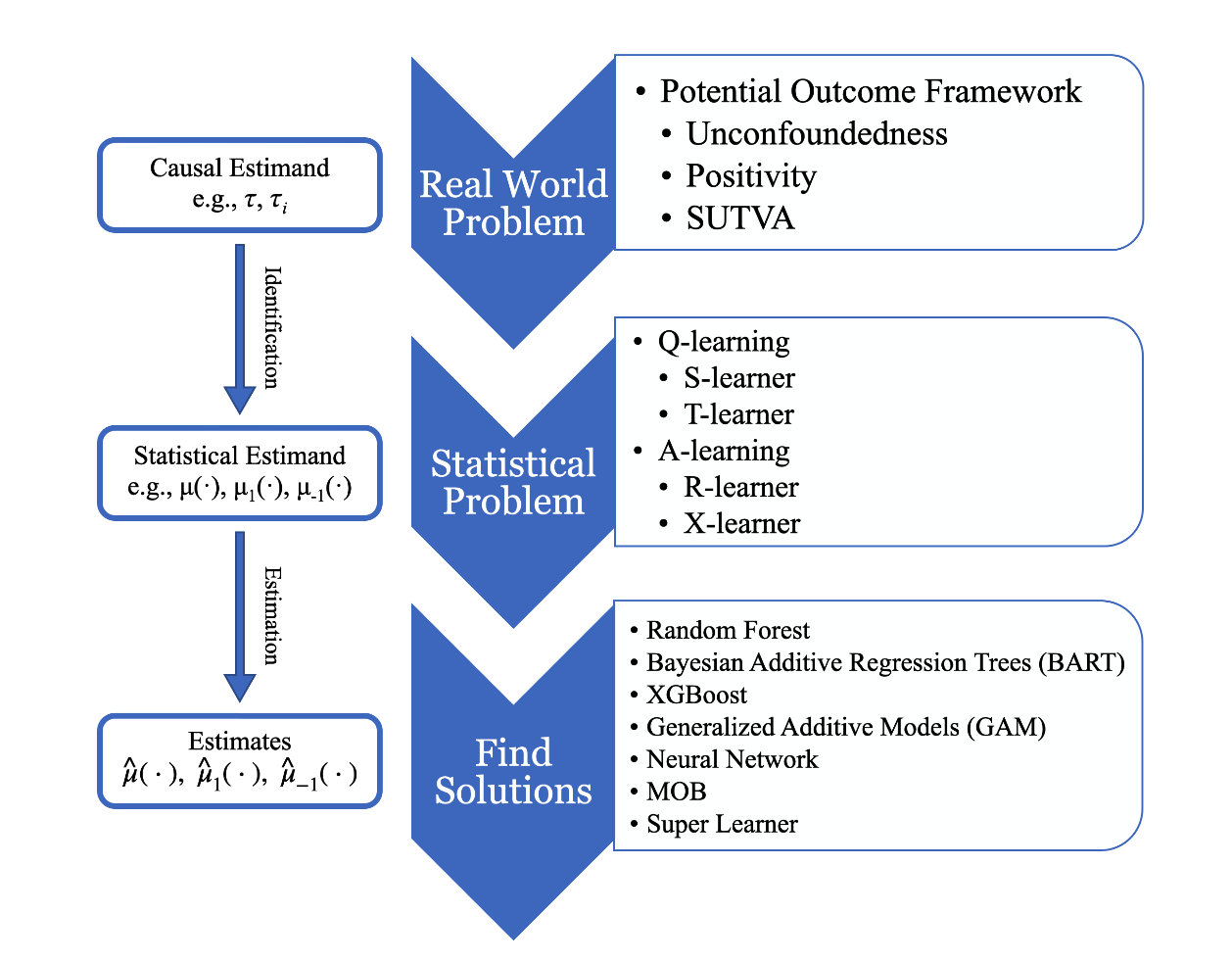
Meta-learners are a simple way to leverage off-the-shelf predictive machine learning methods to estimate conditional average treatment effect (CATE), heterogeneous treatment effect (HTE), and individual treatment effect (ITE). A very general process of doing causal inference is provided in the following flowchart, where there are three main parts:
- Understand the real world question: what is the desired causal estimand/quantity. By potential outcome framework along with several assumptions, the causality could be inferred from the observed data, even without the randomized experiments, which is the gold standard for causal inference.
- In step 2, the original real world problem is transformed to a solvable statistical problem with the assumption made in the first step. Usually, there could be different ways to form the statistical problem with pros and cons. As introduced in Review of Causal Inference: An Overview, there are T-learner and S-learner following the Q-learning approach as well as A-learning-based methods such as R-learner and X-learner.
- After step 2, the statistical problem is built up, so in step 3, the main focus is how to solve them. If viewing in machine learning perspective, step 2 actually yields a problem specific loss function no matter following Q-learning or A-learning approach. The strength of meta-learners is they do not have strong requirements or limitations on the structure of loss function so that any off-the-shelf base learners can be readily fill in to solve. Such as Random Forests (RF) (Breiman 2001), Bayesian Additive Regression Trees (BART) (Chipman et al. 2010), XGBoost (Chen and Guestrin 2016), Generalized Additive Model (GAM) (Hastie and Tibshirani 1986), Neural Network (NN) (Hopfield 1982), Model-Based recursive partitioning (MOB) (Seibold et al. 2016; Zeileis et al. 2008), and Super Learner (SL) (Van der Laan et al. 2007).
2 Multi-arm Treatment Effect Estimation
Most causal inference literature study the treatment effect estimation under two-arm setting; for details, please refer to my another post Review of Causal Inference: An Overview. In this work, we work under a more generalized setting that there could be multiple arms.
2.1 Q-learning
Q-learning gets its name because its objective function plays a role similar to that of the Q or reward function in reinforcement learning (Murphy 2003; Murphy 2005a; Sutton and Barto 2018) and is widely used in estimating optimal dynamic treatment regime (Murphy et al. 2001; Murphy 2003, 2005b; Qian and Murphy 2011; Robins 2004; Schulte et al. 2014). The basic idea of Q-learning is to estimate the conditional response surfaces
In particular, to estimate Q-function, we here consider two approaches, one is Single or S-learner and the other is Two or T-learner method–following the nomenclature in Künzel et al. (2019). Both methods can be easily implemented in either two-arm or multi-arm settings. Notably, here S-, T-, and the following X-, and R-learner, they are merely general frameworks for treatment effect estimation, and in practice, people need to pick proper base learner for their scientific problem, such as Random Forest, BART, XGBoost, and etc., to implement these learners.
2.1.1 S-learner
For S-learner, we estimate a joint function
2.1.2 T-learner
T-learner, on the other hand, estimate
2.2 A-learning
The main difference of advantage-learning or A-learning comparing to Q-learning, is to estimate targeted treatment effect
2.2.1 X-learner
X-learner (Künzel et al. 2019) enjoys the simplicity of T-learner but fixes its data efficiency issue by targeting on the treatment effects rather than the response surfaces. The main procedure for two-treatment setting is (denote two treatments as
- Step 1: Estimate
- Step 2a: Impute ITEs for subjects in
- Step 2b: Fit one model
- Step 3: Combine
For multi-arm setting, we can extend it with the same gist
- Step 1: Estimate
- Step 2a: For any pairwise HTE, for example,
- Step 2b: Fit one model for each imputed HTE which yields two models, denote
- Step 3: Combine
The underlying idea of X-learner is easy to understand: it tries to impute the missing outcomes first via the estimated Q-functions from the T-learner. After obtaining the pseudo ITE
2.2.2 R-learner
R-learner (Nie and Wager 2020) adopts the Robinson’s decomposition (Robinson 1988) to connect the HTE with the observed quantities,
Equation (2.1) is a two-arm version of R-learner. Following (citation), the loss function of R-learner in multi-arm setting is
Similar to X-learner, R-learner is also designed for continuous outcomes due to the limitation of Robinson’s decomposition.
2.2.3 Reference-free R-learner
To deal with the inconsistency recommendation problem in R-learner, the author propose a new method called reference-free R-learner (Zhou et al. 2023) which allows to estimate treatment effect and recommend optimal treatment without specifying a particular reference group. So inconsistency issue is no longer a concern.
The essence of reference-free learner is to reformat the HTE
Then, following Robinson’s idea of decomposition (Robinson 1988), it yields
2.2.4 de-Centralized-Learner
(Author proposed learner which structured for causal inference analysis. Easy to implement and has superior performance comparing to the other meta-learners. For details, please wait for the publication.)
3 Optimal Treatment Recommendation
For S- and T-learner, the optimal treatment given covariate
Since X-learner returns all possible pairwise treatment comparisons,
For R-learner, given a set of estimated HTE,
3.1 Optimal Treatment Regimes
Meta-learner-based approaches find optimal treatment regimes by first estimate the treatment effect and then determine the optimal treatment accordingly. Another branch of methods, however, bypass the needs of estimating treatment effect but directly find the optimal treatment regimes. The reshape the problem as a weighted classification problem and then, many classification tools like SVM and trees can be used to solve the problem (Qian and Murphy 2011; Xu et al. 2015; Zhao et al. 2012; Zhu et al. 2017). Please check the basic idea of these approaches in my another post.
3.1.1 Optimal Dynamic Treatment Regimes (DTR)
In this whole work, we focus on the study with only one time of intervention. However, in practice, multiple-stage interventions may occur and how to determine the optimal sequence of treatments is therefore a natural question. It needs to highlight that subjects cannot be simply treated by the optimal treatment at each stage because the treatment effect may interact with each other and such greedy-algorithm-like idea cannot justify the carry-over effect. Q-learning and A-learning along with the dynamic programming is proposed to learn the optimal DTR from the Sequential, Multiple, Assignment Randomized Trials (SMART) (Lavori and Dawson 2000; Murphy 2005b; Nahum-Shani et al. 2012). If with proper causal assumptions, we can also learn the DTR from the observational studies as well with meta-learners. Since this is a big topic, we illustrate it in my another post in details along with some example codes for better demonstration.